New bacterial strain typing methods in the era of genomics

Microbiology and molecular biotechnology have seen many tremendous advancements over recent years, especially in detecting bacterial infections. One of our previous articles tells you all you need to know about the importance of studying bacterial strains.
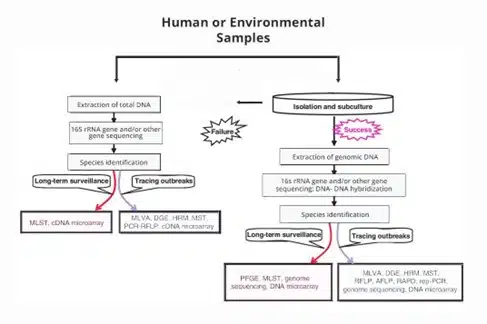
Bacterial strain typing techniques identify bacteria at the strain level and uncover the genetic diversity underlying important phenotypic characteristics. The interspecies diversity of bacteria is the outcome of three genetic processes:
- horizontal gene transfer
- gene recombination
- gene acquisition or loss
At present, there are two principal categories of bacterial strain typing systems: phenotyping and genotyping. Phenotypic characters often reflect observable genetic traits. Genotyping, on the other hand, enables the discrimination of bacterial strains depending on their genetic content.
Bacterial phenotypes are not varied enough to distinguish between closely related strains. Scientists and researchers are, therefore, shifting their focus to genetic or molecular methods for identifying and subtyping bacterial strains. A particular genotyping technique can produce a genetic profile for a strain as distinctive as a fingerprint. Hence, genotyping is also known as DNA fingerprinting.
Helvetica Health Care (HHC) summarises the various current genomic bacterial strain typing methods. Before we delve into these new-generation biotech applications, here is some information on our endeavour to improve bacterial strain typing.
HHC boasts a wide range of INACTIVATED and LIVE ORGANISMS, available in several formats, including CELL EXTRACT, PURIFIED VIRUSES, CELL CULTURES, INACTIVATED CULTURE FLUIDS and DIRECT PELLETED viruses. Our collection of bacterial strains is available as PURIFIED DNA (or NATtrol). Most bacteria are clinical isolates, and the identity of the stock culture is confirmed by 16S ribosomal gene sequencing.
What are the different methods of genotyping bacterial strains?
Currently, there are three main categories of genotyping methods:
1. DNA banding: These methods discriminate the strains under study based on differences in the size of the DNA fragments or bands produced by cleavage of DNA with the help of restriction enzymes (REs) or by amplifying genomic DNA.
2. DNA sequencing: The original nucleotide sequence is generated by DNA sequencing, and this sequence can be used to distinguish between different bacterial strains.
3. DNA hybridisation: By examining the hybridisation of the bacteria’s DNA to known sequence probes, DNA hybridisation-based technologies can distinguish between bacterial strains.
The selective power of genotyping techniques, apart from genome sequencing, depends on the species. Each of these categories includes several methods. Here is a summary of each of them.
1. DNA banding pattern-based methods
Sub-category: Enzymatic restriction
Pulsed-field gel electrophoresis (PFGE): Large DNA molecules can be separated using the electrophoretic method, also known as pulsed-field gel electrophoresis (PFGE) (10 kb–10 Mb).
Restriction fragment length polymorphism (RFLP): RFLP, or restriction fragment length polymorphism, is a technique for measuring the size of restriction fragments that have been separated by standard agarose gel electrophoresis.
Sub-category: DNA amplification
Arbitrarily primed PCR (AP-PCR):
AP-PCR is a PCR variant that uses arbitrary primers to amplify unknown genomic regions at random.
Repetitive sequencing-based PCR (REP-PCR): This method involves distributing numerous copies of a collection of naturally occurring repetitive DNA sequences throughout bacterial genomes.
Multiple-locus variable number tandem repeat analysis (MLVA):
Due to their fast evolution, variable number tandem repeats, also known as VNTRs (tandemly repeated DNA sequences with variable copy numbers and widely distributed in human and bacterial genomes), are an essential source of genetic variation for strain typing. MLVA technique is based on polymorphism analysis of many VNTR sites on chromosomes.
Denaturing gel electrophoresis (DGE):
In DGE, sequence-dependent electrophoresis in polyacrylamide gels are used to separate PCR amplicons of the same length.
High-resolution melting (HRM) analysis:
Similar to DGE, HRM uses melting temperature analysis to distinguish between DNA alleles. Real-time PCR amplification and melting curve analysis are used in HRM.
Sub-category: Enzymatic digestion following DNA amplification
PCR-RFLP:
The limitations of RFLP are overcome by PCR-RFLP, which involves RFLP analysis of a particular locus amplified by PCR. It is a quick, nonradioactive, and easy method to find DNA polymorphism in PCR-RFLP.
Amplified fragment length polymorphism (AFLP):
This technique, which combines the precision of PCR with the accuracy of RE analysis, is very sensitive in identifying DNA polymorphism.
2. DNA sequencing-based methods
The Sanger method:
In this technique, the DNA sequence of a single-stranded template DNA is identified by employing DNA polymerase to create a collection of polynucleotide fragments of various lengths by using dideoxynucleotides that halt the DNA amplification process’ elongation stage.
Pyrosequencing:
Pyrosequencing, a non-electrophoretic sequencing approach based on real-time quantitative measurement of pyrophosphate produced following nucleotide incorporation into a developing DNA chain during DNA synthesis, differs from the traditional Sanger method.
Sub-category: Gene sequencing
Amplification and sequencing of the 16S rRNA gene:
16S rRNA gene is highly conserved among bacteria and other kingdoms because rRNA is essential for the survival of all cells due to its involvement in protein synthesis. Consequently, amplification and sequencing of the 16S rRNA gene are widely used for the identification and phylogenetic classification of prokaryotic species, genera, and families.
A few less conserved genes, particularly those under positive selection, have frequently been employed for bacterial strain typing and, in fewer instances, for species identification. The majority of these genes produce virulence or surface proteins for bacteria. A different set of suitable genes may be selected depending on the species.
Multilocus sequence typing (MLST):
One of the most common genotyping techniques for identifying bacterial strains – MLST uses DNA sequencing to find allelic variants in numerous conserved genes (often seven genes).
16S–23S rRNA gene internal transcribed spacer (ITS):
ITS is a unique genomic region separating the 16S and 23S rRNA genes in prokaryotic microorganisms. The ITS technique comprises the first noncoding sequence utilised for bacterial strain typing.
Multispacer typing (MST):
The MST genotyping approach, based on DNA sequencing, initially used to identify Y. pestis in 2004, uses highly variable intergenic spacers as typing markers.
Sub-category: Genome sequencing
Genome sequencing is the most accurate way of genotyping since it provides access to the entire genetic makeup of bacterial strains. Genome sequencing is done either separately or in combination using the Sanger and pyrosequencing techniques.
3. DNA hybridisation-based methods
DNA macroarrays:
Macroarrays provide a quick, accurate, and affordable genotyping method without the need to buy expensive equipment and be particularly successful in identifying genes involved in antibiotic resistance.
DNA microarrays:
DNA microarrays are carefully prepared microscope slides (chips) that contain an ordered mosaic of sequences that largely or entirely correspond to an organism’s genes. The DNA microarray methodology has rapidly developed into an effective method for examining bacterial transcriptome and genetic diversity.
cDNA microarrays:
A cDNA microarray’s probes, full-length genes amplified by PCR or drawn from cDNA libraries, can be used to find genes linked to serotypes, virulence, antibiotic resistance, epidemics, and host diversity.
Oligonucleotide microarrays:
An oligonucleotide microarray’s probes are brief nucleic acid sequences containing between 30 and 70 nucleotides directly produced or spotted in high density on the solid substrate (up to 1 000 000 oligonucleotides per chip).
HHC strives to expand our collection of viruses and bacteria. Our new products are provided in various formats, designed to meet your research and product development needs. Contact us now to learn about our efforts in improving bacterial strain typing lab applications!